Numpy Random Interface in JAX
Published:
In this blog, we’ll be looking at one of the “sharp bits” of JAX: pseudorandomness, and see if we can’t smooth it out a little. JAX is an accelerated linear algebra library that forms the backbone of some modern python based scientific and machine learning libraries (things like Google’s Gemini were built on JAX). Personally I’m a fan.

This post is mostly an exploration of JAX, and the solutions presented below probably come with a lot of footguns (especially with any sort of sharding/distributed training) and this is meant to be pedagogical rather than practical (in fact, I even prefer the explicit key management of JAX, usually).
So, how does numpy/torch handle randomness and how does JAX differ? The traditional approach of numpy is to have the pseudo-random generator (PRNG) hidden from the user, so each time you call np.random.normal it implicitly handles the generation to be unique. It also allows for you to set a global seed with np.random.seed(int). The JAX approach is to explicitly manage the key which is the driver of the pseudo-random generator (the motivations for JAX’s PRNG design are outlined here). Historically, these impure functions have not meshed well with JAX’s design. While JAX’s design has a lot of benefits, sometimes you just want a random number and don’t want to think about the key (case in point: https://docs.jax.dev/en/latest/pytrees.html#example-of-jax-tree-map-with-ml-model-parameters).
However, now that array refs are part of the exposed API, we can make things a little more numpy-like. Now, we all know what arrays are, but what is an array ref? That is beyond the scope of this blog (a clever way of saying: I’m not really sure myself), so let’s just use the physicist explanation of spin and say an array ref is like a C pointer except it’s not a pointer, and it’s not a first class citizen1.
With array refs, it’s quite straightforward to make a more numpy style array interface (in fact, there’s a JEP for this exact thing). We can just make a stateful ref counter, and increment that for stateful randomness. To be specific, consider the following:
import jax
from jax import numpy as jnp
from dataclasses import dataclass, field
@jax.tree_util.register_dataclass
@dataclass
class RNG:
key: jax.Array
counter: jax.Ref
def __init__(self, seed):
self.key = jax.random.key(seed)
self.counter = jax.new_ref(jnp.array(0, dtype=jnp.int32))
def normal(self, shape = (), dtype = None):
self.counter[...] = self.counter[...] + 1
key = jax.random.fold_in(self.key, self.counter[...])
return jax.random.normal(key, shape=shape, dtype=dtype)
def set_seed(self, seed):
self.counter[...] = jnp.array(seed)
rng = RNG(0)
Then, when we use it in a package, we might see something like,
from jox import rng as jr
jr.set_seed(0)
weights = jr.normal((10, 10))
However, right off the bat, this comes with a problem. We can’t vmap over it. If you try, you see a Exception: performing a set/swap operation with vmapped value on an unbatched array reference of type Ref{int32[]}. Move the array reference to be an argument to the vmapped function? error. The reason for that is the ref is being closed over, and so when the vmap-ing happens, JAX doesn’t know what the ref should become (since many operations in parallel are happening to the ref). Vmap is like 1/3 of the JAX transforms, so this is a bit of a problem. There’s (at least) two possible solutions. First, we can make the actual PRNG object be passed around and allow for splitting the object so it can also be vmap-ed over (if the ref is an argument of the function, you can vmap over refs). This is what the current JEP does. But the second route is to double down and embrace the numpy interface (the whole point of this was to avoid managing an explicit state as a user, key or otherwise). We can accomplish this second route because we know something specific about the outcome of our ref: it doesn’t depend on the inputs. The usual problem with closed over refs is if you do some operation on the inputs to the vmap and the ref depends on them, it’s unclear what the output should be. However, we know our ref doesn’t depend on the values of the array (we can just increment the ref by the batch size, or even just by 1).
One way to accomplish this second approach is with a custom vmap rule. However, since we aren’t explicitly vmap-ing over the jr.normal, the custom vmap rule won’t necessarily trigger. So we have to make our implementation recognize when it is inside of vmap. This can be done using axis names, via the following:
@jax.custom_batching.custom_vmap
def _normal(counter, key, batch_idx, shape, dtype):
counter[...] = counter[...] + 1
key = jax.random.fold_in(key, counter[...])
return jax.random.normal(key, shape=shape, dtype=dtype)
@_normal.def_vmap
def _normal_vmap_rule(axis_size, in_batched, counter, key, batch_idx, shape, dtype):
# Add by batch size (could just be 1?)
counter[...] += axis_size
k = jax.random.fold_in(key, counter[...])
result = jax.random.normal(k, shape=(axis_size, *shape), dtype=dtype)
return result, True
@jax.tree_util.register_dataclass
@dataclass
class RNG:
key: jax.Array
counter: jax.Ref
axis: str = field(metadata=dict(static=True))
def __init__(self, seed, axis="i"):
self.key = jax.random.key(seed)
self.counter = jax.new_ref(jnp.array(0, dtype=jnp.int32))
self.axis = axis
def normal(self, shape = (), dtype = None):
try:
batch_idx = jax.lax.axis_index(self.axis)
except:
batch_idx = jnp.array(0)
return _normal(self.counter, self.key, batch_idx, shape, dtype)
def set_seed(self, seed):
self.counter[...] = jnp.array(seed)
rng = RNG(0)
This allows for vmap support, albeit not perfect since the user has to specify the axis name (also this isn’t robust to nested vmaps but could be made to be)2. However, this is only at the vmap level and is a minor change (that could also be optimized at the interface). If they don’t specify it, the code will run, but all the values will be identical (silent PRNG failures are always preferable to raising errors right?).
def f(x):
return jr.normal() + x
jax.vmap(f, axis_name="i")(jnp.ones(10)) # works!

Technically, we have one more hurdle to go through. If you set the shape (even outside of a jit), you will see an error like TypeError: Shapes must be 1D sequences of concrete values of integer type, got (JitTracer<~int32[]>, JitTracer<~int32[]>). This is because the custom vmap code is immediately traced for a jaxpr, which means the static integers get promoted to tracers. It is not clear to me whether this is necessary, and the fix is to add something like static arguments (like custom vjp) or whether there is a simpler solution3. However, we can skirt around this for now by also capturing the shapes:
def normal(self, shape = (), dtype = None):
try:
batch_idx = jax.lax.axis_index(self.axis)
except:
batch_idx = jnp.array(0)
@jax.custom_batching.custom_vmap
def _normal(counter, key, batch_idx):
counter[...] = counter[...] + 1
key = jax.random.fold_in(key, counter[...])
return jax.random.normal(key, shape=shape, dtype=dtype)
@_normal.def_vmap
def _normal_vmap_rule(axis_size, in_batched, counter, key, batch_idx):
# Add by batch size
counter[...] += axis_size
k = jax.random.fold_in(key, counter[...])
result = jax.random.normal(k, shape=(axis_size, *shape), dtype=dtype)
return result, True
return _normal(self.counter, self.key, batch_idx)
To prove this approach actually works, here is a pure JAX VAE, with absolutely no jax.random.key and no jax.random.split. We can visualize the latent space in the same way as https://keras.io/examples/generative/vae/,
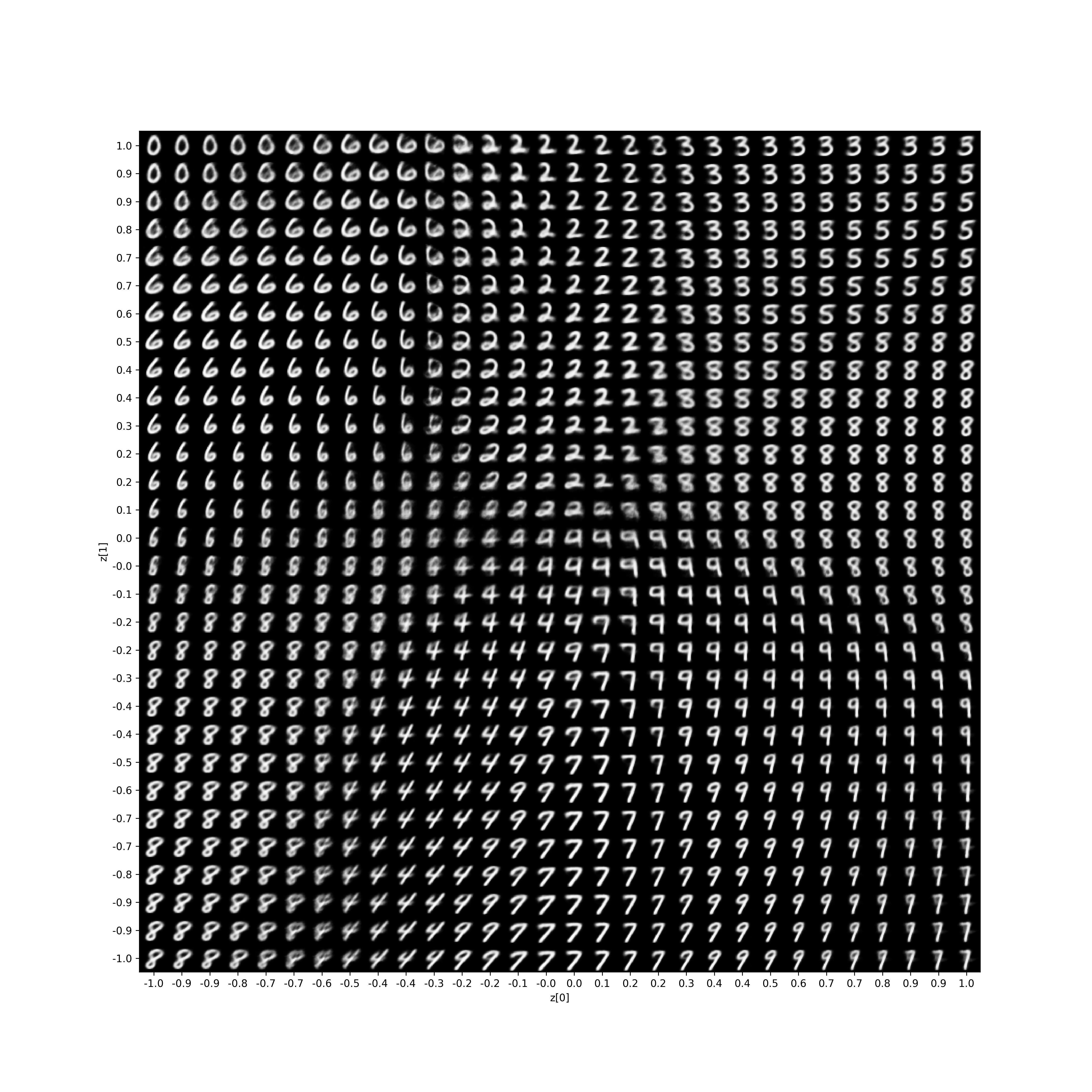
Full Training Code
import jax
import jax.numpy as jnp
from jox import rng as jr
import optax
import matplotlib.pyplot as plt
import numpy as np
from torchvision import datasets
train = datasets.MNIST(root='./data', train=True, download=True)
test = datasets.MNIST(root='./data', train=False, download=True)
x_all = np.concatenate([train.data.numpy(), test.data.numpy()], axis=0)
x_all = x_all.astype(np.float32) / 255.0
x_all = x_all.reshape(-1, 1, 28, 28)
batch_size = 100
num_samples = (len(x_all) // batch_size) * batch_size
x = jnp.array(x_all[:num_samples].reshape(-1, batch_size, 1, 28, 28))
print(f"Data shape: {x.shape}")
plt.matshow(x[0][0].reshape((28, 28)), cmap='gray')
plt.show()
def init_linear(in_features, out_features):
stddev = jnp.sqrt(2.0 / (in_features + out_features))
w = jr.normal((in_features, out_features)) * stddev
b = jnp.zeros(out_features)
return {'w': w, 'b': b}
def linear(params, x):
return x @ params['w'] + params['b']
def init_vae(input_dim=784, hidden_dims=[512, 256], latent_dim=2):
params = {'encoder': [], 'decoder': [], 'z_mean': None, 'z_log_var': None}
key_idx = 0
enc_dims = [input_dim] + hidden_dims
for i in range(len(enc_dims) - 1):
params['encoder'].append(init_linear(enc_dims[i], enc_dims[i+1]))
key_idx += 1
params['z_mean'] = init_linear(hidden_dims[-1], latent_dim)
key_idx += 1
params['z_log_var'] = init_linear( hidden_dims[-1], latent_dim)
key_idx += 1
dec_dims = [latent_dim] + hidden_dims[::-1] + [input_dim]
for i in range(len(dec_dims) - 1):
params['decoder'].append(init_linear(dec_dims[i], dec_dims[i+1]))
key_idx += 1
return params
def encode(params, x):
h = x.flatten()
for layer_params in params['encoder']:
h = jax.nn.relu(linear(layer_params, h))
z_mean = linear(params['z_mean'], h)
z_log_var = linear(params['z_log_var'], h)
z = z_mean + jnp.exp(0.5 * z_log_var) * jr.normal(shape=z_mean.shape)
return z_mean, z_log_var, z
def decode(params, z):
h = z
for layer_params in params['decoder'][:-1]:
h = jax.nn.relu(linear(layer_params, h))
logits = linear(params['decoder'][-1], h)
return logits.reshape(1, 28, 28)
def forward(params, x):
z_mean, z_log_var, z = encode(params, x)
logits = decode(params, z)
return z_mean, z_log_var, logits
def binary_cross_entropy(labels, logits):
# https://github.com/google/flax/blob/main/examples/vae/train.py
logits = jax.nn.log_sigmoid(logits)
return -jnp.sum(labels * logits + (1.0 - labels) * jnp.log(-jnp.expm1(logits)))
def vae_loss_single(logits, z_mean, z_log_var, data):
recon_loss = jnp.sum(binary_cross_entropy(data, logits))
kl_loss = -0.5 * jnp.sum(1 + z_log_var - jnp.square(z_mean) - jnp.exp(z_log_var))
return recon_loss + kl_loss
def loss_fn(params, data):
def single_loss(x):
z_mean, z_log_var, logits = forward(params, x)
return vae_loss_single(logits, z_mean, z_log_var, x)
losses = jax.vmap(single_loss, axis_name="i")(data)
return jnp.mean(losses)
@jax.jit
def step(params, opt_state, data):
loss_value, grads = jax.value_and_grad(loss_fn)(params, data)
updates, opt_state = optimizer.update(grads, opt_state, params)
params = optax.apply_updates(params, updates)
return params, opt_state, loss_value
params = init_vae(input_dim=784, hidden_dims=[512, 256], latent_dim=2)
optimizer = optax.adam(learning_rate=0.001)
opt_state = optimizer.init(params)
losses = []
for i in range(100):
if i % 10 == 0:
print(i)
batch_losses = []
for batch in x:
params, opt_state, l = step(params, opt_state, batch)
batch_losses.append(l)
losses.append(np.mean(np.array(batch_losses)))
plt.plot(losses)
plt.xlabel('Epoch')
plt.ylabel('Loss')
plt.show()
def plot_latent_space(params, n=30, figsize=15):
digit_size = 28
scale = 1.0
figure = np.zeros((digit_size * n, digit_size * n))
grid_x = jnp.linspace(-scale, scale, n)
grid_y = jnp.linspace(-scale, scale, n)[::-1]
for i, yi in enumerate(grid_y):
for j, xi in enumerate(grid_x):
z_sample = jnp.array([xi, yi])
x_decoded = jax.nn.sigmoid(decode(params, z_sample))
digit = x_decoded[0].reshape(digit_size, digit_size)
figure[
i * digit_size : (i + 1) * digit_size,
j * digit_size : (j + 1) * digit_size,
] = digit
plt.figure(figsize=(figsize, figsize))
start_range = digit_size // 2
end_range = n * digit_size + start_range
pixel_range = jnp.arange(start_range, end_range, digit_size)
sample_range_x = [float(round(i, 1)) for i in grid_x.tolist()]
sample_range_y = [float(round(i, 1)) for i in grid_y.tolist()]
plt.xticks(pixel_range, sample_range_x)
plt.yticks(pixel_range, sample_range_y)
plt.xlabel("z[0]")
plt.ylabel("z[1]")
plt.imshow(figure, cmap="Greys_r")
plt.show()
plot_latent_space(params)
Changelog
- December 15, 2025: Published initial version.
- February 2, 2026: Fixed some typos.
Footnotes
How do array refs interact with the C/C++ FFI interface? I’m not sure, but that’s an interesting question. ↩
Another possible solution might be to make a custom primitive, but I haven’t given that as much thought. ↩
This might be fixable in JAX, I will update this blog as this JAX issue progresses https://github.com/jax-ml/jax/issues/33943. ↩
